We aim at designing novel processes, techniques and technology for the analysis, design, development and integration of complex information systems, exploring synergies in related topics.
The research interests of IDSS span three key areas. The Information Systems Engineering area focuses on Enterprise architecture and engineering, Business process management, Requirements engineering, Model-driven engineering, and Knowledge engineering.
The Data Science and Engineering area focuses on Machine learning, Algorithms and complexity, Data profiling, cleaning and integration, Data analytics, Information retrieval, and Spatial data science. Finally, the Algorithms Engineering area has a focus on Algorithms and data structures, Succinct data structures, Combinatorial optimization, and Computational science.
Projects

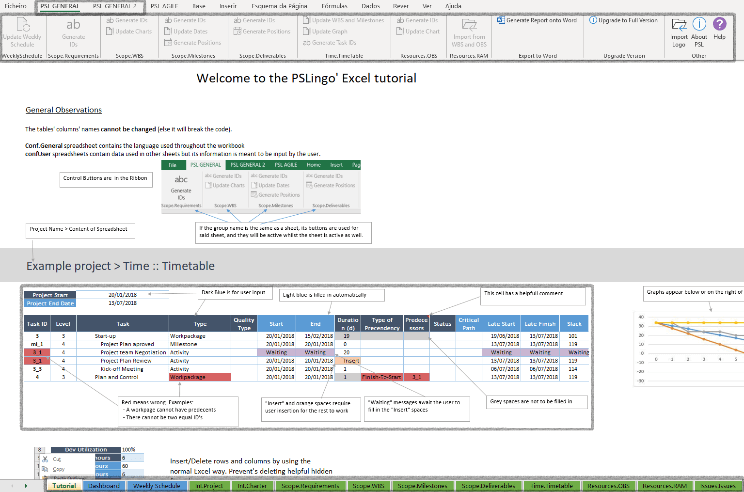
Research tool: PSL Excel Tutorial
Complex Network
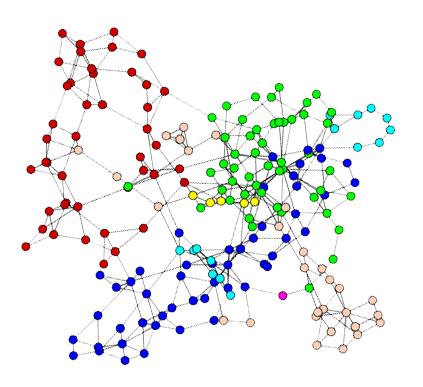
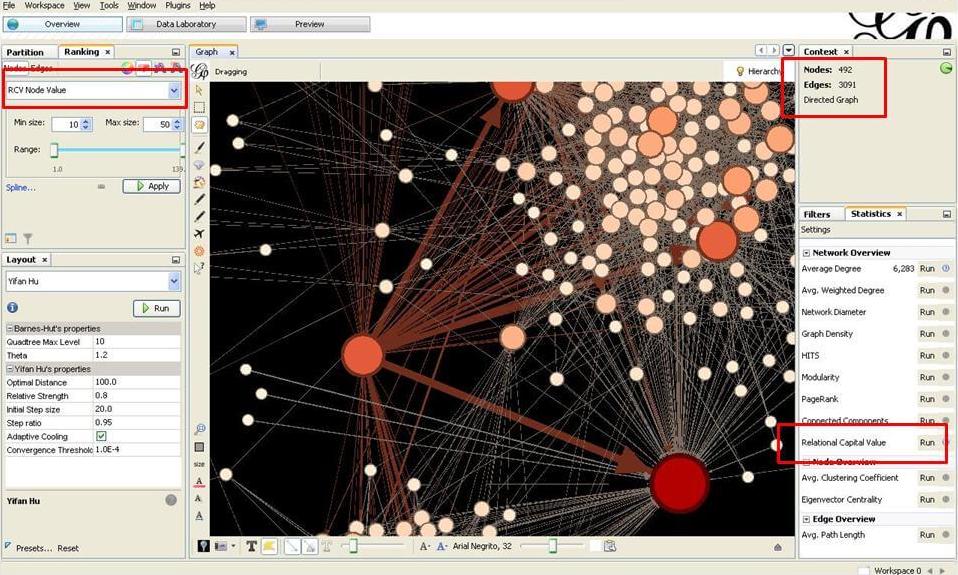
Gephi’s Plugin (Social Networks)
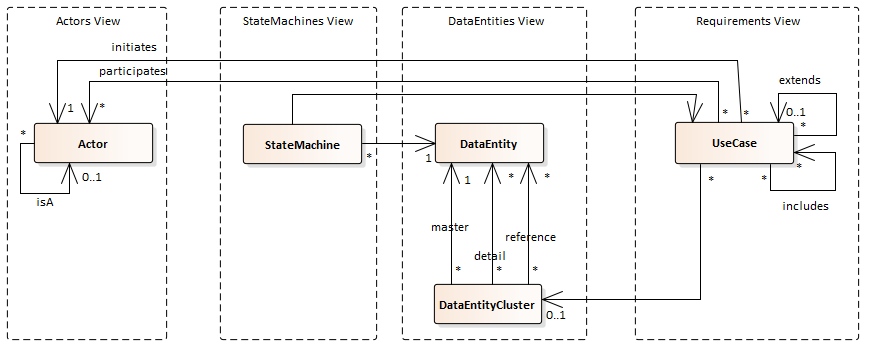
RSL (partial) Metamodel